Before you begin
You can process and report UNIFI® analysis results using Watson LIMSTM. You can import a Watson LIMS-generated analytical run into UNIFI software and use it as a sample list for an analysis. You can export the analysis results back into the analytical run, to enable processing and reporting of those results in Watson LIMS. Before you can perform these actions, you must configure UNIFI software to recognize Watson LIMS.
Prerequisites:
- Thermo Fisher personnel installed Watson LIMS on a computer other than the UNIFI server.
- If the application/database server is in a different domain than that of the LIMS server, ensure that the date and time-stamps are synchronized to a time server. In a Windows® Server 2012 R2-based domain, the domain controllers are automatically configured as time servers for the domain.
- You or a Waters Service Engineer can access the Watson LIMS Integration ManagerTM Admin Client and Watson LIMS Integration Manager Service Manager applications.See also: These Thermo Scientific guides and their associated release notes:
- Integration Manager 3.0 Administration Guide, Revision A
- Integration Manager 3.0 Watson Agent Configuration Guide, Revision A
- Integration Manager 3.0 Forms Agent Configuration Guide, Revision A
- Integration Manager 3.0 Installation and Configuration Guide, Revision A
- Integration Manager 3.0 Web Service Agent Configuration Guide, Revision A
Configure Watson LIMS to work with UNIFI software as follows:
- Configure a Watson agent as instructed in the Integration Manager 3.0 Watson Agent Configuration Guide, Revision A.
- Configure a Forms Agent as instructed in the Integration Manager 3.0 Forms Agent Configuration Guide, Revision A.
- Connect the import IPO from the Watson agent to an export IPO on the forms agent by means of an interface and interface map.
- Record the external interface URL for the Forms Agent.
Result: UNIFI interface software reads the LIML sent by Watson from the URL and IPO specified in the Forms Agent.
- Using the Forms and Watson agents previously created, create an interface and interface map from an IMPORT IPO on the forms agent to an Export IPO on the Watson agent. These interface maps transfer data pushed into the forms agent URL (to the Unique Import IPO) to the Watson agent where it is queued for import.
- Add instrument interface to Watson LIMS.
- Update instrument parameter file.
Configuring Watson agent
Watson agents connect the Integration Manager and the Watson LIMS via the Watson IM Gateway.
You must perform these tasks to create and configure a Watson agent:
- Create a new Watson agent, using the IM Service Manager as specified in the Watson Agent Configuration Guide, Revision A.
- Configure the Watson agent, using the IM Administration as specified in the Watson Agent Configuration Guide, Revision A.
Configuring Forms agent
You must configure the Forms agent, as specified in the Integration Manager 3.0 Forms Agent Configuration Guide, Revision A.
The Forms agent contains the external service URL.
Requirement: Record the external service URL. You must specify this value when you configure LIMS in UNIFI software.
You must add a new integration point to export analytical assays into UNIFI software.
To configure Forms agent to work with UNIFI software:
- Open Integration Manager and then click System Configuration > Configure Agents.
- On the Integration Manager Form Agents tab, select FormsAgent1, and then click Attributes.Result: The FormsAgent1 Attributes dialog box lists the External Service URL value.

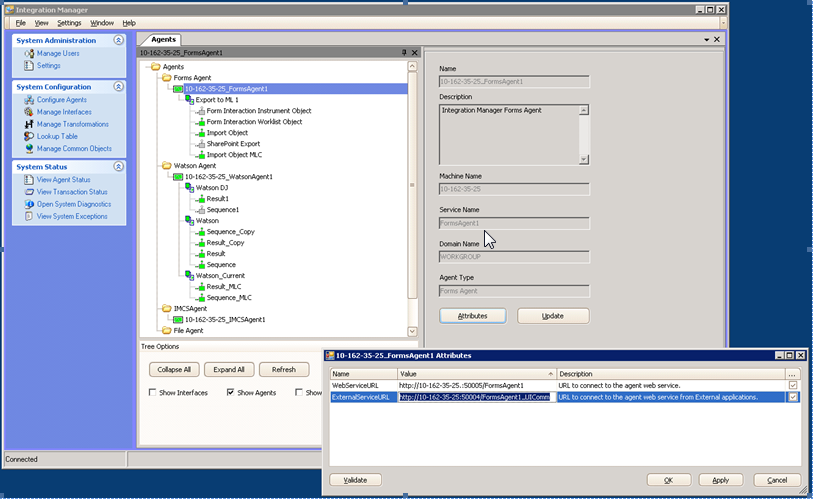
- Add a new Integration Point Object by performing these steps:
- Right-click Forms Agent 1 > New Integration Point.
- In the New dialog box, enter “Export to UNIFI” for the name, an optional description, and then click OK.
- For these items, keep the default values as specified in the Forms Agent Configuration Guide, Revision A.
- Forms Interaction Worklist Object and its attributes.Tip: When you configure LIMS in UNIFI software, you must enter the Integration Point Object Code (WORKLIST_TO_FORM) in the Sample Lists IP Object Code field of the Custom Tab in the Configuration Detail page.
- Import Object and its attributes.Tip: When you configure LIMS in UNIFI software, you must enter the Import Object name (Import Object) in the Results Object field of the Custom Tab in the Configuration Detail page.
Creating an interface for the Integration Manager
After the Watson agent and Forms agent are configured correctly, you must connect the import IPO from the Watson Agent to an export IPO on the Forms agent by means of an interface and interface map. You must create and map an Integration Manager interface between Watson LIMS and UNIFI software and UNIFI software and Watson LIMS.
See also: Thermo Scientific Integration Manager 3.0 Administration Guide, Revision A
To create and map an IM interface between Watson LIMS to UNIFI software:
- Open Integration Manager, and then click System Configuration > Manage Interfaces.
- From the Interfaces page, right-click an empty area of the tree view pane, and then select Add New Interface.
- In the New Interface wizard page, enter “Watson to UNIFI” for the name, leave the description blank, and then click Next.
- Select Integration Points, and then click Next:
- For Integration point 1 select “Watson” on agent WatsonAgent1
- For Integration point 2, select “Export to UNIFI” on agent FormsAgent1
- Specify “30 seconds” as the Polling Interval and Unit and then click Next.
- Enter the following values, click Add Map, and then click Finish.
Interface map values:
| Name |
Watson to UNIFI_Map1 |
| Description |
Leave blank |
| Interface map code |
Leave blank |
| IP 1 Object |
Sequence |
| IP 2 Object |
Form Interaction Worklist Object |
To create and map an IM interface between UNIFI software and Watson LIMS:
- Open Integration Manager, and then click System Configuration > Manage Interfaces.
- From the Interfaces page, right-click an empty area of the tree view pane, and then select Add New Interface.
- In the New Interface wizard page, enter “UNIFI to Watson” for the name, leave the description blank, and then click Next.
- Select Integration Points and then click Next:
- For Integration point 1 select “Export to UNIFI” on agent FormsAgent1.
- For Integration point 2, select “Watson” on agent WatsonAgent1.
- Specify “30 seconds” as the Polling Interval and Unit, and then click Next.
- Enter the following values, click Add Map, and then click Finish.
Interface map values:
| Name |
UNIFI to Watson_Map |
| Description |
Leave blank |
| Interface map code |
Leave blank |
| IP 1 Object |
Form Interaction Worklist Object |
| IP 2 Object |
Sequence |
Adding instrument interface to Watson LIMS
You must create an instrument interface to export your analytical run assays to use in software. You can make the sample tray of the instrument interface specific to your instrument system. You can create multiple instrument interfaces for other sample trays and titer plates.
To add a new instrument interface:
- Log on to Watson LIMS.
- Click Options > Instrument Interface.
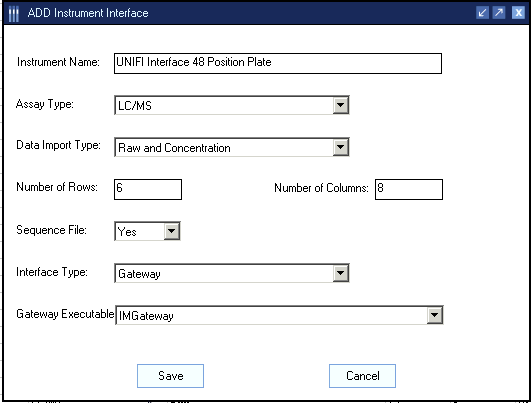
- In the Add Instrument Interface dialog box, enter the following values, and then click Save.
Instrument interface values:
| Instrument Name |
Enter a name that describes your instrument configuration and include the word “UNIFI” in the name.
Example: UNIFI Interface 48 Position Plate. |
| Assay Type |
LC/MS |
| Data Import Type |
Raw and Concentration |
| Number of Rows |
Enter the number of rows in a sample tray or titer plate.
Example: Enter 6 for a 48-position vial tray. |
| Number of Columns |
Enter the number of columns in a sample tray or titer plate.
Example: Enter 8 for a 48-position vial tray. |
| Sequence File |
Yes |
| Interface type |
Gateway |
| Gateway Executable |
IM Gateway |
Add instrument interface dialog box:

Updating the instrument parameter file
When you create a new instrument interface, you must update the IM Instrument Parameters file to include the name of the newly created instrument interface (UNIFI Interface 48 Position Plate in the previous example) and the corresponding WatsonAgentURL (Integration Manager connection). If you do not update this file, an error message informs you that the new instrument interface is missing and that the Watson agent cannot read the Watson Agent URL.
To update the IM Instrument Parameters File:
- Browse to Program Files\Thermo\Watson directory, and open the IMInstrumentParameters.XML file in Notepad.
- Add these lines to this file:<Instrument InstrumentName=”UNIFI Interface 48 Position Plate”><!–where “UNIFI Interface 48 Position Plate” equals the name of the newly created instrument interface–><Parameter ParameterName=”WatsonAgentURL” ParameterRequired=”Y”>
<Value ParameterValue=”soap.tcp://10-162-35-25.:50006/WatsonAgent1″/>
</Parameter>
</Instrument>
Creating new LIMS configuration in UNIFI software
Requirement: Your role must include the Create and modify LIMS configuration permission.
To create a new LIMS configuration:
- Log on to software.
- Click Administration > LIMS configuration.
- On the LIMS Configuration page, click Create.
- On the General Tab of the Configuration Detail page, enter a name and description for the LIMS configuration.Restriction: Do not select the Disable Import check box. When you want to remove the LIMS configuration, you can select this check box to prevent users from importing sample information into UNIFI software.
- On the Custom Tab of the Configuration Detail page, enter the connection information.Requirement: The connection information in the Custom tab must match the settings specified in the Forms Agent page of Watson LIMS Integration Manager application.
- Type the Forms Agent Web Service URL. The FormsAgent1 Attributes dialog box in Integration Manager identifies the External Service URL.
- In the Sample Lists IP Object Code field, type WORKLIST_TO_FORM.
- In the Results IP Object field, type Import Object.

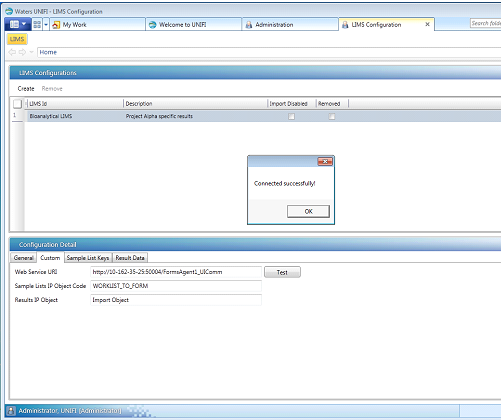
- Click Test.Result: If UNIFI software can connect to LIMS, the “Connected successfully!” message appears. If the connection is unsuccessful, ensure that the Web service URL matches the External Service URL specified in the Forms Agent page of Watson LIMS Integration Manager application.
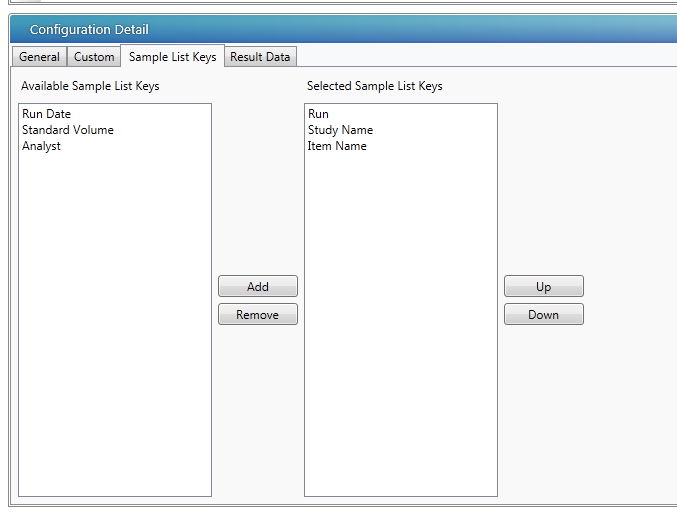
- In the Sample List Keys tab of the Configuration Detail page, select the common, analytical-run assay field names. These sample list keys relate to metadata exported from the LIMS analytical run, which are used to automatically generate UNIFI analysis name. Use the Up and Down buttons to change the order of the sample keys. (The order of selected sample keys defines the order of list and is used as a name for the new analysis.)

- On the Result Data tab of the Configuration Detail page, select Response, or both Response and Observed RT, specifying analysis results for export to LIMS using this configuration.

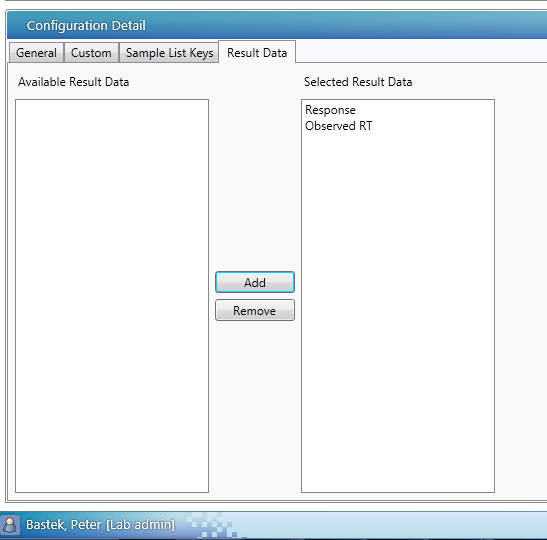
- Click Save.
Creating an analysis for LIMS
After you configure the LIMS interface, you can create an analysis to be used by LIMS.
To create an analysis for LIMS:
- From Watson LIMS, click File > Select Project.
- Click Analytical > Analytical Run Actions.
- Select a Run ID, and then click the Sequence File button.

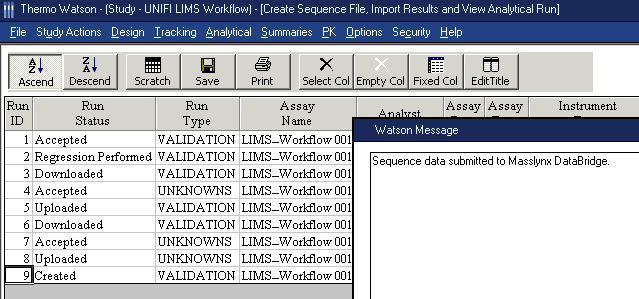 Result: This message appears: “Preparing to create MassLynx DataBridge Sequence Data”.Tip: “MassLnyx Databridge Sequence” is a configurable name that is specified on the Import IPO page in the Watson Agent.
Result: This message appears: “Preparing to create MassLynx DataBridge Sequence Data”.Tip: “MassLnyx Databridge Sequence” is a configurable name that is specified on the Import IPO page in the Watson Agent.
- In the Value column of the instrument Parameters dialog box, select Watson <<Sequence>>, and then click OK.
- When the “Sequence data submitted to Masslynx DataBridge” message appears in the Watson Message dialog box, click OK.
Result: A UNIFI-compatible sample list is available to pick from the “Use sample list from LIMS” section of the Create Analysis wizard in UNIFI software.
Creating an analysis in UNIFI
You can now create an analysis in UNIFI software to use with LIMS.
To create an analysis to use with LIMS:
- Log on to UNIFI software.
- On the My Work tab, click Create > Analysis.
- On the Create Analysis wizard page, click “Create analysis by acquiring new data”, and then click Next.
- In the Create Analysis wizard on the page “What sample list and analysis method do you want to use?”, click “Use sample list from LIMS”, and then select the LIMS configuration you created previously created.


- In the LIMS Sample Lists dialog box, select the sample list that you created in LIMS, and then click OK.

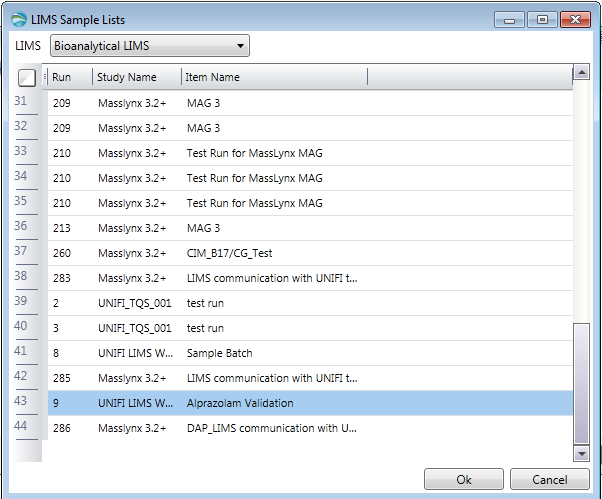
Result: Doing so ensures the LIMS-created sample list is used as the sample list for the current analysis.
Exporting analysis to LIMS
When a LIMS-generated analysis is completed, you can export it back into the original LIMS Analytical Run Assay for processing.
To export the LIMS analysis into Watson LIMS:
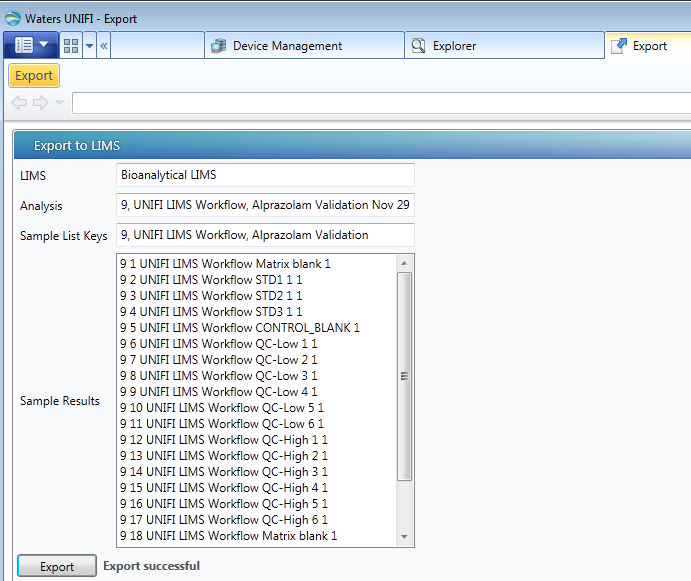
- Log on to UNIFI software.
- Open Explorer, select the analysis, and then right-click Export to > LIMS.
- In the Export to LIMS page, click Export.Result: The “Export successful” message appears at the bottom of the page.

- Open Watson LIMS.
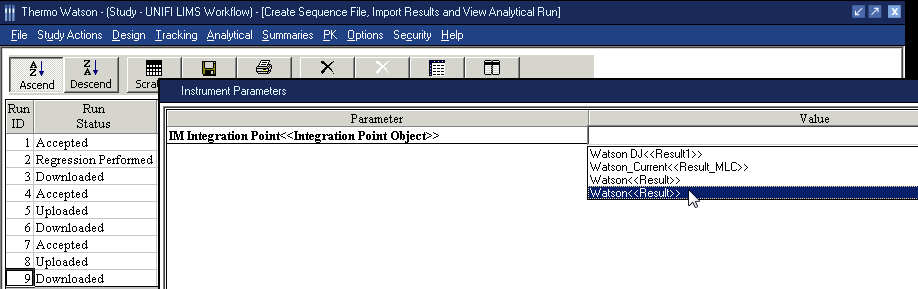
- Select the Run ID of the analytical run assay that matches the analysis results being exported, and then click the Import button.
- In the Instrument Parameters dialog box, click Watson <<Results>> in the Value column.

- In the second Instrument Parameters dialog box, select the analysis result in the Value column.

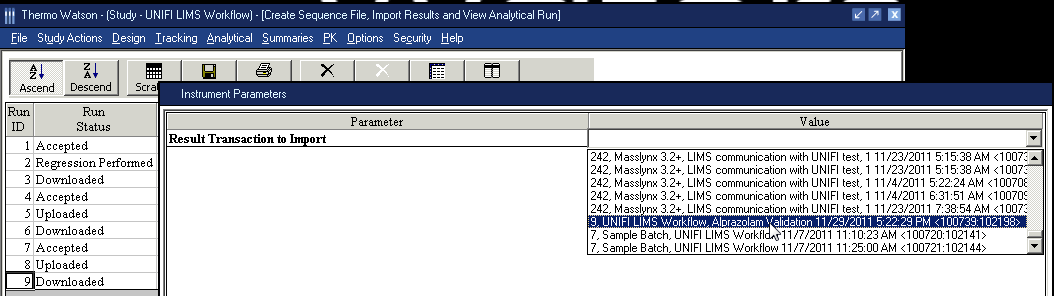 Requirement: You can import the UNIFI analysis only to the matching Analytical Run Assay. The date and time that the analysis was exported from UNIFI software must match that the date and time the result was imported into Watson LIMS.Tip: Match the sample list key information to the analytical run assay information.
Requirement: You can import the UNIFI analysis only to the matching Analytical Run Assay. The date and time that the analysis was exported from UNIFI software must match that the date and time the result was imported into Watson LIMS.Tip: Match the sample list key information to the analytical run assay information.
- Click OK.
- To find the correct imported results in Watson LIMS, search the Import File Name column in the Analytical Run Actions page.Results: The imported data appear in LIMS.Tip: NaN indicates that the value is not a number and that, consequently, no result was found.
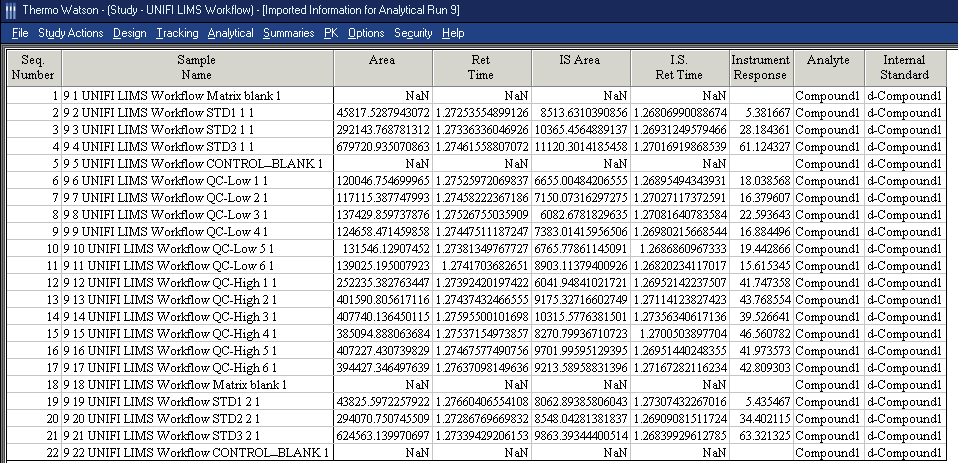

Viewing exported results
When you export an analysis from UNIFI software and import it into Watson LIMS, you must match the UNIFI analysis to the analytical run assay in LIMS. You do so by comparing the date and time stamps that report when the analysis was exported from UNIFI and the result imported into LIMS. To easily identify the date the analysis was exported, create a specialized view in UNIFI’s Explorer window.
To create a specialized view in Explorer:
- Log on to UNIFI software.
- Open Explorer, click the View list on the toolbar, and then click Select View > Analyses and Reports.
- Right-click any column heading in the table, and then click Add Column.
- In the Add Columns dialog box, perform the following tasks, and then click OK:
- Expand Common Fields
- Click Last result export
- Click Add
- From the View list on the toolbar, click Current View > Save As.
- In the Save View dialog box, enter a description for the view, make it public or private, and then click OK.

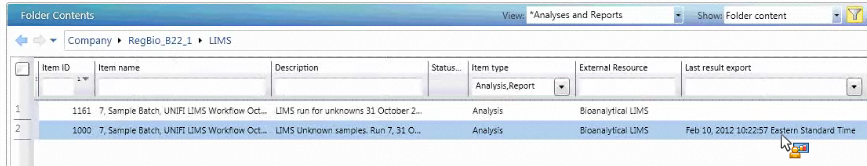
Tip: You can move the “Last result export” column by dragging and dropping it to the desired location in the Folders Contents table. By default, the column is positioned as the last right-hand column.
Result: The “Last result export” column displays the date and time that the analysis was exported from UNIFI software into LIMS.